Triovis - Using trio-information for depth-based read filtering
One of the most important steps in SNP discovery is to remove false positive, for example by filtering on read depth for each SNP. Unfortunately, the cut-offs that one uses (typically: remove SNPs with coverage lower than 3 or higher than 1,200) are based on the 1,000 Genome Project data analysis and might not be ideal for the data at hand. When sequencing trios (father, mother and offspring), it is however possible to include the trio-information at the filtering stage. For example, many SNPs in the filtered data where both parents are reference/reference and the offspring is alternative/alternative, indicates that the there are issues with the cut-off values used. The Triovis tool provides live feedback of what the effects of these different cut-offs are on “possible” and “impossible” inheritance patterns.
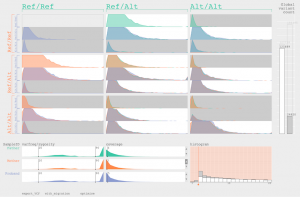
Reference: Sakai, Ryo, Alejandro Sifrim, Andrew Vande Moere, and Jan Aerts. 2013. “TrioVis: A Visualisation Approach for Filtering Genomic Variants of Parent-Child Trios.” Bioinformatics (Oxford, England) (May 8): 2–3.