Hierarchical Boundary Coefficient Clustering
HBCC is a clustering algorithm is inspired by the work of Peng et al. [4]. Their CDC algorithm detects regions that separate clusters by quantifying how spread out points’ neighbors are in feature space. The intuition for this process is as follows: (1) points within a clusters have near neighbors on all sides, and (2) points in-between clusters or on the edge of a cluster predominantly have neighbors in the direction of the cluster’s core.
The same intuition was previously used by Vandaele et al. [2] to
quantify how close points are to the boundary of the data’s manifold.
They used their boundary coefficient to extract manifold skeletons.
HBCC combines Vandaele et al. [2]’s boundary coefficient with HDBSCAN’s
[1], [3] principled density-based cluster selection strategies. This
approach removes CDC’s ratio parameter. Instead a clustering
hierarchy is evaluated over all boundary coefficient values and
clusters are selected from the hierarchy.
The implementation relies on McInnes’ fast_hdbscan and its support for lensed clustering introduced for Bot et al. [5] to detect branches.
The HBCC algorithm works as follows:
Compute k nearest neighbors and minimum spanning tree [3].
Compute n-hop distances from nearest neighbors [2].
Compute the boundary coefficient for each point [2].
Compute minimum spanning tree from boundary coefficient weighted knn–mst graph union [5].
Compute HDBSCAN cluster hierarchy and selection [1].
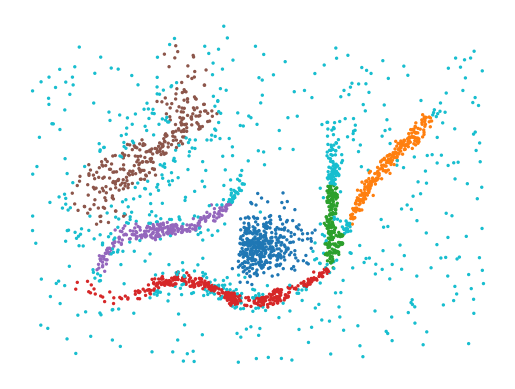
How to use HBCC
import numpy as np
import matplotlib.pyplot as plt
from fast_hbcc import HBCC
data = np.load('data/flareable/flared_clusterable_data.npy')
c = HBCC(
num_hops=4,
min_samples=10,
min_cluster_size=100,
cluster_selection_method="leaf",
).fit(data)
plt.scatter(*data.T, c=c.labels_ % 10, s=2, cmap='tab10', vmin=0, vmax=9)
plt.axis("off")
plt.show()
Citing
Citing information not available yet.
Licensing
The fast_hbcc package has a 2-Clause BSD license.