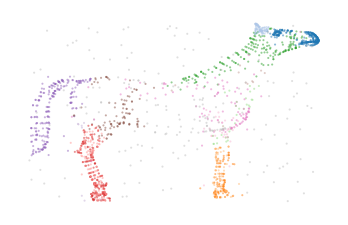
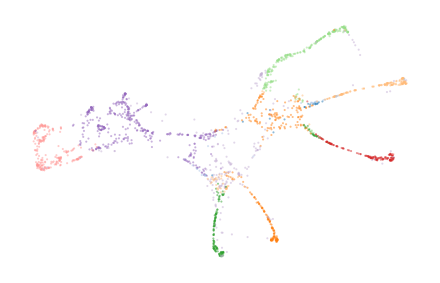
Bi-Persistence Clustering on 3D Horse-shaped Mesh Reconstruction Data
This notebook demonstrates the BPSCAN clustering algorithm on a 3D horse-shaped mesh reconstruction data set. The data was obtained from a STAD repository. The meshes were originally created or adapted for a paper by Robert W. Sumner and Jovan Popovic (2004). They can be downloaded and are described in more detail on their website.
[11]:
%load_ext autoreload
%autoreload 2
[1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from umap import UMAP
from biperscan import BPSCAN
from lib.plotting import *
palette = configure_matplotlib()
Uniformly distributed noise points are added to make the clustering task more difficult.
[2]:
def noise_from(values, num_samples):
min, max = values.min(), values.max()
range = max - min
offset = 0.1 * range
return np.random.uniform(min - offset, max + offset, num_samples)
horse = pd.read_csv("data/horse/horse.csv").sample(frac=0.2)
num_noise_samples = horse.shape[0] // 10
df = pd.concat(
(
horse[["x", "y", "z"]],
pd.DataFrame(
dict(
x=noise_from(horse.x, num_noise_samples),
y=noise_from(horse.y, num_noise_samples),
z=noise_from(horse.z, num_noise_samples),
)
),
)
)
The BPSCAN algorithm requires a bit of tuning to extract the branches. In particular, performance appears better when min_samples is smaller than min_cluster_size. distance_fraction is used to filter out the noise points. Values up to \(0.1\) appear to work quite well in general.
[3]:
c = BPSCAN(min_samples=15, min_cluster_size=100, distance_fraction=0.05).fit(df)
Visualizing a side-view of the dataset makes it difficult to see whether the legs are correctly clustered. So, we also show a 2D UMAP projection that separates the legs and tail.
[4]:
p = UMAP(n_neighbors=50, repulsion_strength=0.1).fit(df.values)
c:\Users\jelme\micromamba\envs\work\lib\site-packages\sklearn\utils\deprecation.py:151: FutureWarning: 'force_all_finite' was renamed to 'ensure_all_finite' in 1.6 and will be removed in 1.8.
warnings.warn(
[5]:
sized_fig(0.5)
plt.scatter(df.z, df.y, s=1, c=c.lens_values_, cmap="viridis")
plt.subplots_adjust(0, 0, 1, 1)
plt.axis("off")
sized_fig(0.5)
plt.scatter(*p.embedding_.T, s=1, c=c.lens_values_, cmap="viridis")
plt.subplots_adjust(0, 0, 1, 1)
plt.axis("off")
plt.show()
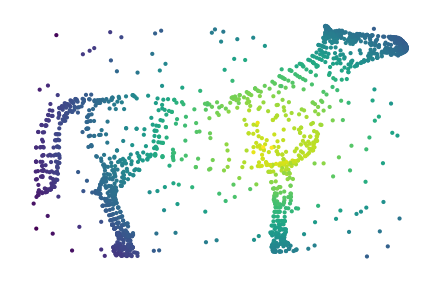
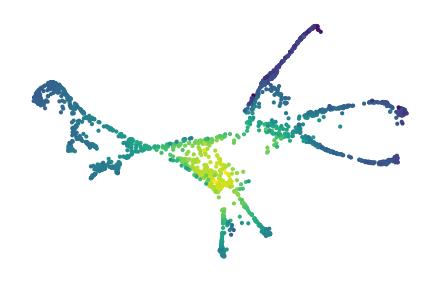
26 merges are detected corresponding to the horse’s legs, head, and tail.
[6]:
sized_fig(0.5, aspect=0.8)
merges = c.merges_
merges.plot_merges(df.z.to_numpy(), df.y.to_numpy(), s=0.33, title_y=0.6, arrowsize=3, linewidth=0.5)
plt.subplots_adjust(0, 0, 1, 0.975, wspace=0)
plt.axis("off")
plt.show()
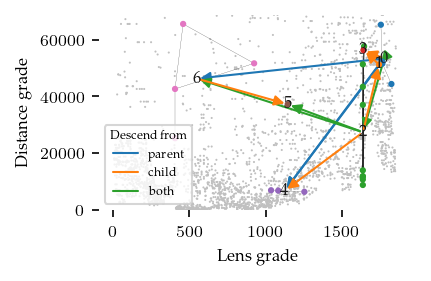
Combining the (near) duplicate merges results in 8 simplified merges that exist at several stages in the bi-filtration:
[7]:
sized_fig(0.5)
c.simplified_merges_.plot_persistence_areas()
plt.show()
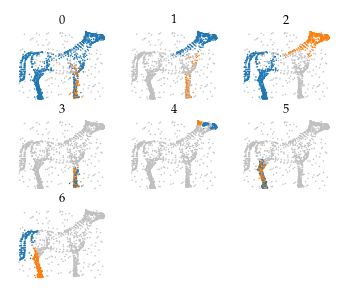
The simplified merges still describe the legs, tail, head, and (depending on the parameters setting) even ears and nose are detected as branches.
[8]:
sized_fig(0.5, 0.8)
c.simplified_merges_.plot_merges(df.z.to_numpy(), df.y.to_numpy(), s=0.5)
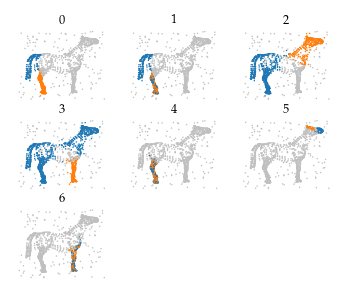
The cluster labels extracted from these merges segment the data set fairly well:
[9]:
scatter_kwargs = dict(s=1, alpha=0.5, linewidths=0, edgecolor="none")
colors = plt.cm.tab20.colors
color_kwargs = dict(
cmap=ListedColormap(["silver"] + [colors[i] for i in range(20)]), vmin=-1, vmax=19
)
[10]:
sized_fig(0.5)
plt.scatter(df.z, df.y, c=c.labels_ % 10, **color_kwargs, **scatter_kwargs)
plt.subplots_adjust(0, 0, 1, 1)
plt.axis("off")
sized_fig(0.5)
plt.scatter(*p.embedding_.T, c=c.labels_ % 10, **color_kwargs, **scatter_kwargs)
plt.subplots_adjust(0, 0, 1, 1)
plt.axis("off")
plt.show()